Montanari et al., 2021
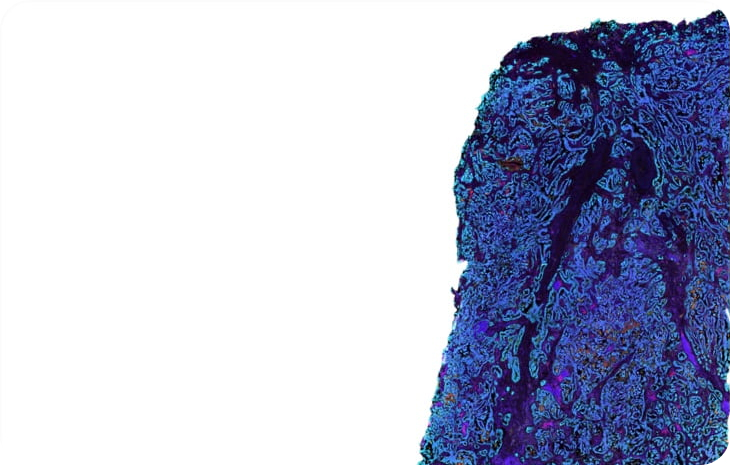
Phenotyping and imaging of the tumor microenvironment using FlexVUE™
Relevant for
Biotech, AMCs, Exploratory Research
Background and aims: Chronic HBV is clinically defined in 4 phases by a combination of serum HBV DNA levels, HBeAg status and ALT: immunotolerant (IT), immune-active (IA), inactive carrier (IC) and HBeAg-negative hepatitis (ENEG). Immune and virological differences between phases as detected in blood have proven useful but are insufficient to appreciate the interrelation between the immune system and the virus since immune dynamics in blood and liver differ. Furthermore, the inflammatory response in the liver and parenchymal cells cannot be fully captured in blood.
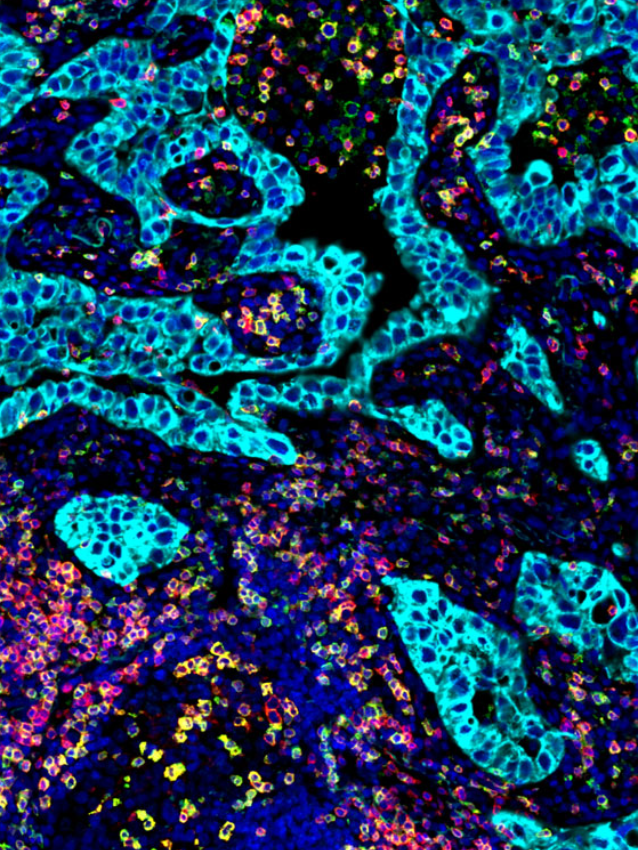
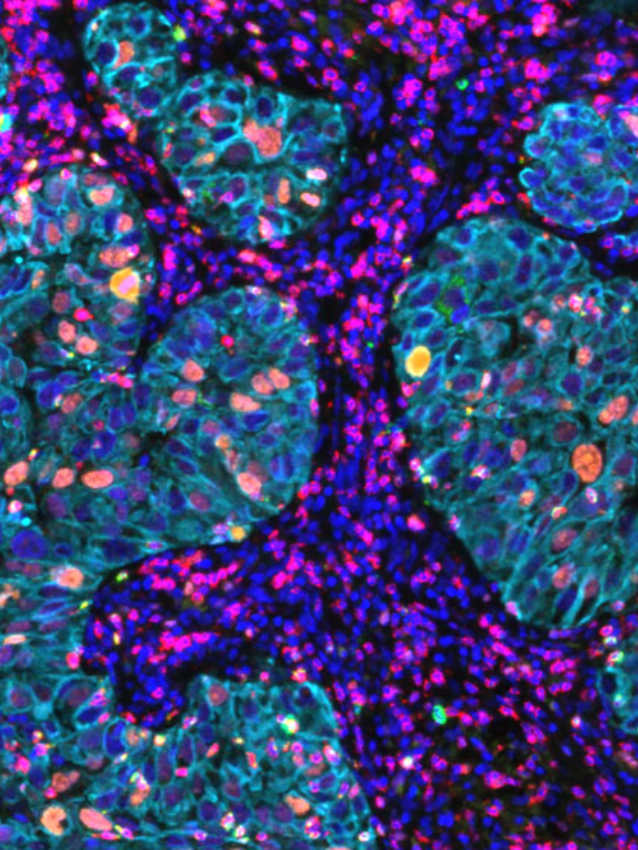
Methods: Immunological composition and transcriptional profiles of FFPE core needle liver-biopsies in chronic HBV phases vs healthy were evaluated by multiplex immunofluorescence and RNA-Seq (n = 37 and 78, respectively).
Results: Irrespective of the phase-specific serological profiles, increased immune-gene expression and frequency was observed in chronic HBV compared to healthy livers. Greater transcriptomic de-regulation was seen in IA and ENEG (172 vs 243 DEGs) than in IT and IC (13 vs 35 DEGs) livers. ISG, immune-activation and exhaustion genes (ICOS, CTLA4, PDCD1) together with chemokine genes (CXCL10, CXCL9) were significantly induced in IA and ENEG livers. Moreover, distinct immune profiles associated with ALT elevation, and a more accentuated immune-exhaustion profile (CTLA4, TOX, SLAMF6, FOXP3) found in ENEG and set it apart from IA phase (LGALS9, PDCD1). Interestingly, all HBV phases showed downregulation of metabolic pathways vs healthy livers (fatty and bile acid metabolism). Finally, we found increased leukocyte infiltrate correlated with serum ALT, but not with HBV DNA or viral proteins.
Conclusion: Our comprehensive multi-parametric analysis of human livers revealed distinct inflammatory profiles and pronounced differences in intrahepatic gene profiles across all chronic HBV phases in comparison to healthy liver.
Summary: Immunological studies on chronic HBV remains largely restricted to assessment of peripheral responses due to the limited access to the site of infection, the liver. In this study, we comprehensively analyzed livers from a well-defined cohort of chronic HBV patients and uninfected controls with state-of-the-art techniques and evaluated the differences in gene expression profiles and inflammation characteristics across distinct disease phases of chronic HBV patients.
What is the problem?
Evaluate the differences in gene expression profiles and inflammation
When we consider digital pathology applications most of our current knowledge has been gleaned from Immunohistochemistry (IHC). While this technique has helped identify targets and cell types as part of the tumor microenvironment (TME), the data has been limited by laborious protocols to only 1-2 markers. Its become clear that IHC is not sufficient enough to generate the necessary information from the comprehensive landscape of the TME and indeed to date, one has to superimpose multiple slides of data to achieve the level of multiplexing required.
Current clinical stratification for NSCLC immunotherapy regimens requires a positive measurement of tumor cell surface PD-L1 expression and a derived tumor proportion score (TPS) score. However co-localization analysis of the immune checkpoint marker PD-L1 and other immune markers ideally requires the detection of multiple markers on the same tissue slide. The goal of the study was to perform a comparison of DNA-barcoding mIF using Ultivue’s InSituPlex technology and conventional IHC to assess PD-L1 status and other immune cell types in NSCLC tumors
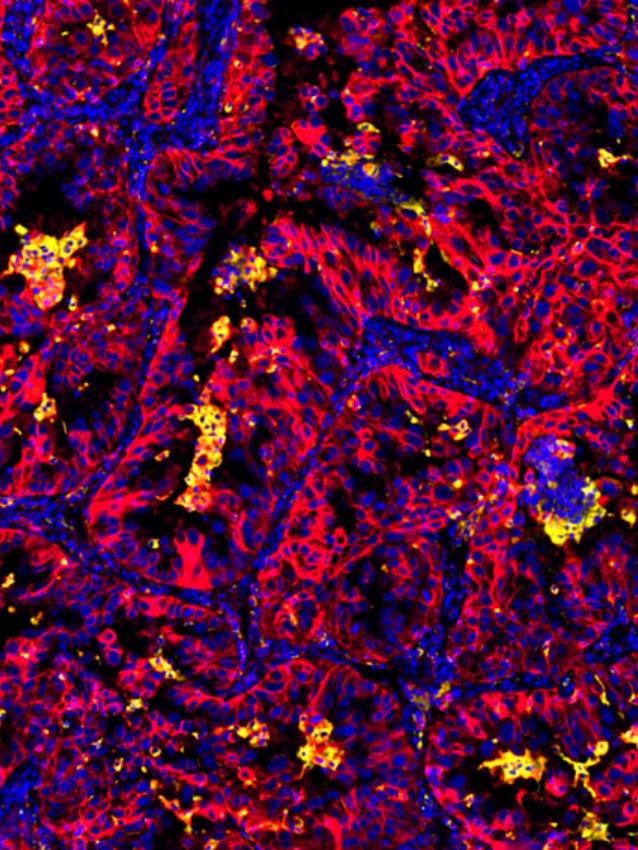
Multiplex immunofluorescence (mIF) and the use of digital image analysis is useful for the quantitative assessment of PD-L1 and other immune cells and is highly consistent with traditional single marker IHC staining results. However,the implementation of mIF and digital image analysis provides a unique insight into cell subsets, specific phenotype co-localization and spatial relations of immune checkpoints (like PD-L1) to tumor cells.
Results
Findings from the study demonstrated a high correlation in immune cell densities and TPS between IHC and mIF.
Traditional IHC of PD-L1 was performed using the PD-L1 kit (PD-L1 22C3, Dako) and multiplexed PD-L1 together with expression of CD68, CD8 and PanCK/Sox10 was enabled by the Ultivue FixVUE PD-L1 pre-optimized kit.
Findings from the study demonstrated a high correlation in immune cell densities and TPS between IHC and mIF. However greater detail and insights into cellular identity and biomarker expression were achieved using mIF. Specifically, simultaneous detection of both PD-L1+ and cytokeratin epithelial cell marker+ (CK+) using ISP technology enabled differentiation of PD-L1+ tumor cells from PD-L1+ T cells and PD-L1+ macrophages.
Further, detection of PD-L1+CK+ and CK+ cells permitted calculation of a quantitative rather than a qualitative TPS. Studies also showed that the calculation of TPS using mIF compared favorably to the TPS calculated using IHC and pathologist review.